The mystery behind HIV And the CD4+ T cells

By Minnie Zhu
Human immunodeficiency virus (HIV) is an infection that targets the body’s immune system. The virus targets specifically the CD4 receptors on CD4+ T cells, a type of T-helper white blood cell containing the CD4 receptors in the lymph nodes.
By targeting the white blood cells and eventually destroying them, the HIV virus weakens the body’s immune system by decreasing white blood cell levels, causing the body to be more prone to coinfections and certain HIV-related cancers. Patients with AIDS are much more susceptible to developing tuberculosis disease, HBV and HCV (Hepatitis B&C), HPV (Human papillomavirus), and CMV (cytomegalovirus). This results in HIV having high case-fatality rates. Even today, HIV has caused roughly 40.4 million deaths globally since the pandemic, and patients with AIDS have a near 100% death rate within 10 years without treatment. Yet, HIV remains to be a global issue with no complete cure for the infection.
What is HIV?
Examining from a cellular level of HIV infection, lets zoom in on the Virus.
HIV is a retrovirus: a family of RNA viruses that produces a complementary DNA copy of its viral RNA. It integrates this DNA into the host cell’s DNA and allows HIV viruses to be produced during replication cycles, eventually taking over the cell.
The diagram below examines how the HIV virus infects and takes over the host cell:
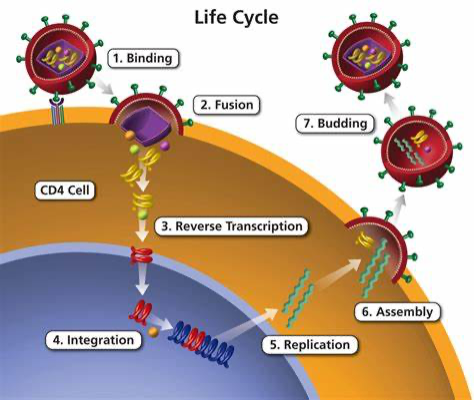
1. Initially, HIV enters the cell due to the recognition of gp120 glycoprotein by the cd4 receptors on the T cells. This allows CD4 T cells to change from a close to an open configuration, allowing HIV to then fuse with the cell.
2. Once bound, the glycoprotein gp41 on HIV mediates a dynamin-dependent fusion with the host endosomal membrane. And the core of the HIV- the nucleocapsid is then released into the cell
(*there is still a partial coating on the nucleocapsid).
3. Once in the cell, the virus undergoes reverse transcription: opposite to transcription, the virus reverses the process and produces DNA from RNA with the use of an enzyme Reverse transcriptase.
4. The final transcribed DNA is covalently attached to the host cell’s DNA through the aid of virus-integration enzymes– encoded integrase (IN) protein. The HIV integration machinery must also interact with many additional host factors for integration, including nuclear trafficking and pore proteins during nuclear entry, histones during initial target capture, and DNA repair proteins during the completion of the DNA joining steps.
5. After integration, the cell would express the DNA gene as it replicates, allowing viral DNA to also be reproduced.
6. HIV would then be assembled and released through the plasma membrane in the process of budding.
7. Eventually, after a few replications, HIV would destroy the host cell, causing the cell to terminate.
CD4-gp120 protein-virus complex :
The CD4-gp120 protein-virus complex plays a pivotal role in virus binding and fusion.
Here is a basic of how the protein and virus fuse:
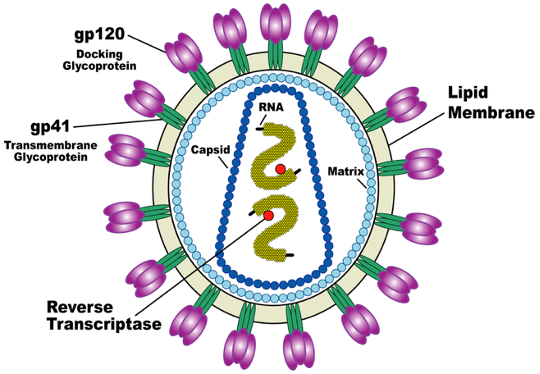
On the outer membrane of HIV, the virus is surrounded by a lipid envelope, which fuses with the host cell’s membrane during infection. This entire process is mediated by the HIV-1 Env glycoprotein complex embedded in the lipid membrane. Glycoprotein is a group of linked amino acids connected to carbohydrates which often function for cell signalling and cell-cell recognition.
The env trimer protein complex (gp120/gp413) contains 1 gp120 surface docking glycoprotein to attach to CD4 receptors and 3 gp41 trimetric transmembrane glycoprotein for structural foundation and mediates fusion of the membranes. This complex surrounds the nucleocapsid of the virus and coats the outside of the virus. This env (gp120/gp413) trimer protein complex is the sole virus-specific protein present on the viral membrane.
Phe43 in gp120 glycoprotein of HIV
During infection, HIV binds to the CD4 protein tag which mainly exists on the surface of helper T (Th) cells; CCR5 and CXCR4 which are secondary receptors are also crucial to binding as these receptors control the availability of T cells to bind to the virus and stabilizes the binding. The specific binding site on the CD4+ T cells that gp120 binds to is the phe43 cavity, a conserved region between the inner and outer region of the CD4 receptor.
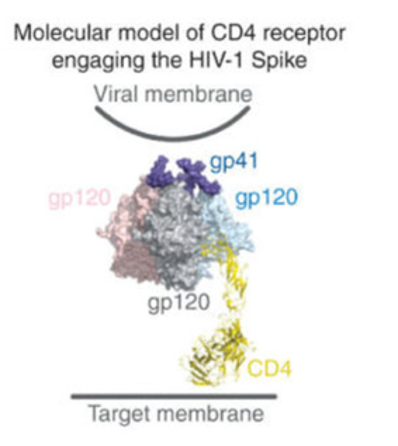
In the unliganded form, the env trimer on HIV virus exists in a structurally constrained ‘closed’ conformation. Only engagement with the receptor, gp120 undergoes a series of structural rearrangements resulting in the sequential opening of the env trimer into an asymmetric intermediate and ending in an “open” conformation. By opening up the receptor, the HIV finishes its binding to the host cell as gp41 then meditates membrane fusion and reverse transcription begins, taking control over the host cells.
Reference:
1. Moir, Susan et al. “Pathogenic mechanisms of HIV disease.” Annual review of pathology vol. 6 (2011): 223-48. doi:10.1146/annurev-pathol-011110-130254
2. Camuto, Alan. “HIV and Co-Infections.” AmfAR, the Foundation for AIDS Research, 29 Mar. 2022, www.amfar.org/treat-asia/hiv-and-co-infections/.
3. Boulougoura, Afroditi, and Irini Sereti. “HIV infection and immune activation: the role of coinfections.” Current opinion in HIV and AIDS vol. 11,2 (2016): 191-200. doi:10.1097/COH.0000000000000241
4. Nnakenyi, Ifeyinwa Dorothy et al. “Prevalence of hepatitis B and C virus co-infection in HIV positive patients attending a health institution in southeast Nigeria.” African health sciences vol. 20,2 (2020): 579-586. doi:10.4314/ahs.v20i2.5
5. “HIV and AIDS Epidemic Global Overview.” HIV.gov, 2022, www.hiv.gov/federal-response/pepfar-global-aids/global-hiv-aids-overview/.
6. Shrestha, Lok Bahadur et al. “Co-infection of Hepatitis B and Hepatitis C among HIV-infected patients: A cross-sectional study from tertiary care hospital of eastern Nepal.” PloS one vol. 17,3 e0264791. 3 Mar. 2022, doi:10.1371/journal.pone.0264791
7. World. “HIV and AIDS.” Who.int, World Health Organization: WHO, 13 July 2023, www.who.int/news-room/fact-sheets/detail/hiv-aids?gclid=CjwKCAjwivemBhBhEiwAJxNWN5CWd2XKc-
8. Rambaut, Andrew et al. “The causes and consequences of HIV evolution.” Nature reviews. Genetics vol. 5,1 (2004): 52-61. doi:10.1038/nrg1246
9. Ribeiro, Ruy M et al. “Estimation of the initial viral growth rate and basic reproductive number during acute HIV-1 infection.” Journal of virology vol. 84,12 (2010): 6096-102. doi:10.1128/JVI.00127-10
10. Coffin, John, and Ronald Swanstrom. “HIV pathogenesis: dynamics and genetics of viral populations and infected cells.” Cold Spring Harbor perspectives in medicine vol. 3,1 a012526. 1 Jan. 2013, doi:10.1101/cshperspect.a012526
11. Stammers, D. K., and Jingshan Ren. “Structure of Retrovirus Particles (Retroviridae).” Elsevier EBooks, Jan. 2021, pp. 352–61, https://doi.org/10.1016/b978-0-12-814515-9.00109-0
12. Schlub, Timothy E et al. “Fifteen to twenty percent of HIV substitution mutations are associated with recombination.” Journal of virology vol. 88,7 (2014): 3837-49. doi:10.1128/JVI.03136-13
13. Balzer, Deb. “Forty Years of HIV/AIDS: Will the Epidemic End? - Mayo Clinic News Network.” Mayo Clinic News Network, 14 June 2021,
14. “1. VIROLOGY of HUMAN IMMUNODEFICIENCY VIRUS.” Aids.gov.hk, 2023, www.aids.gov.hk/pdf/g190htm/01.htm.
15. Jérémie Prévost, et al. “The HIV-1 Env Gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.” MBio, vol. 11, no. 3, American Society for Microbiology, June 2020, https://doi.org/10.1128/mbio.00280-20
16. Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associ- ated with control of viremia in primary human immunodefi- ciency virus type 1 infection. J Virol 1994;68:6103-10.
17. AllenTM,O’ConnorDH,JingP,DzurisJL,MothéBR,Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 2000;407:386-90.
18. “HIV Spikes.” Ucsf.edu, 2023, www.cgl.ucsf.edu/chimera/data/hiv09/hiv-demo.html.
19. Carlon-Andres, I., Malinauskas, T. & Padilla-Parra, S. Structure dynamics of HIV-1 Env trimers on native virions engaged with living T cells. Commun Biol 4, 1228 (2021). https://doi.org/10.1038/s42003-021-02658-1
20. Zwick, M., Saphire, E. & Burton, D. gp41: HIV's shy protein. Nat Med 10, 133–134 (2004). https://doi.org/10.1038/nm0204-133
21. Mercier, L. (2023). Human immunodeficiency virus 1 ~ ViralZone. Expasy.org. https://viralzone.expasy.org/7
22. Craigie, R., & Bushman, F. D. (2012). HIV DNA Integration. Cold Spring Harbor Perspectives in Medicine, 2(7), a006890–a006890. https://doi.org/10.1101/cshperspect.a006890